What is it about?
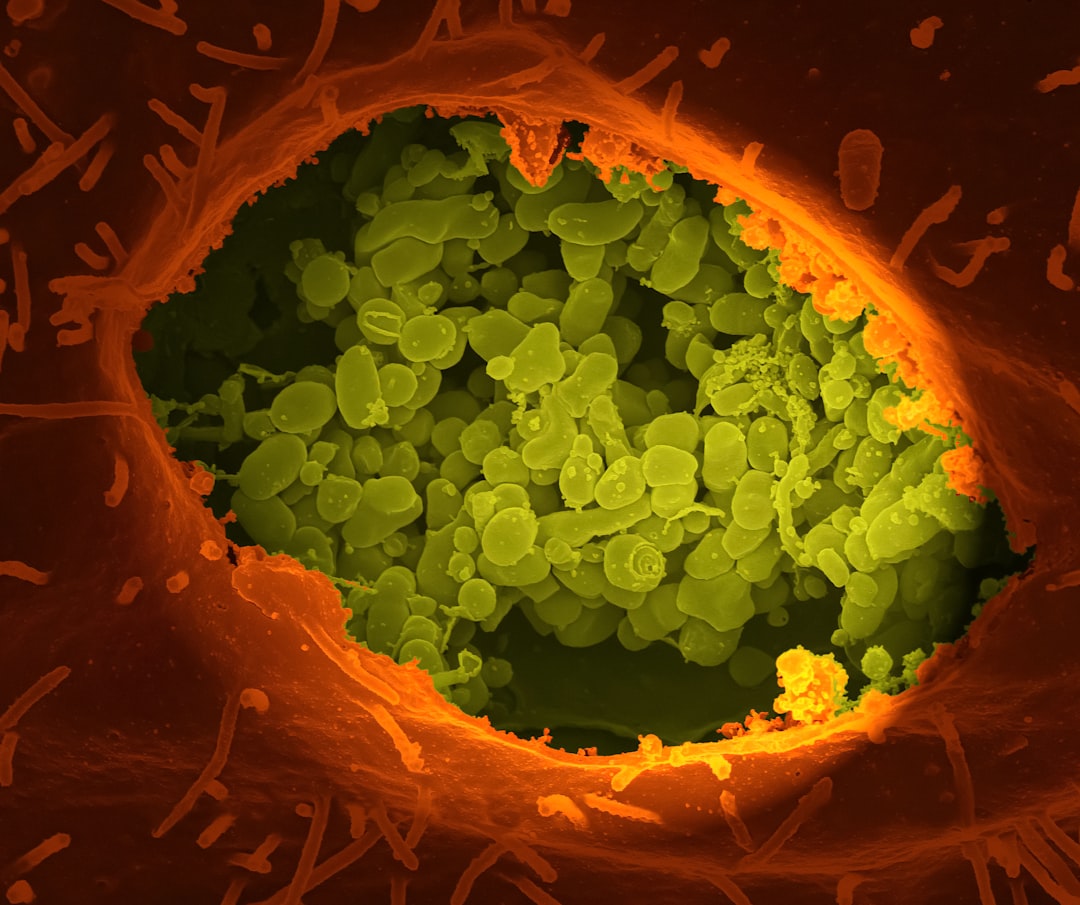
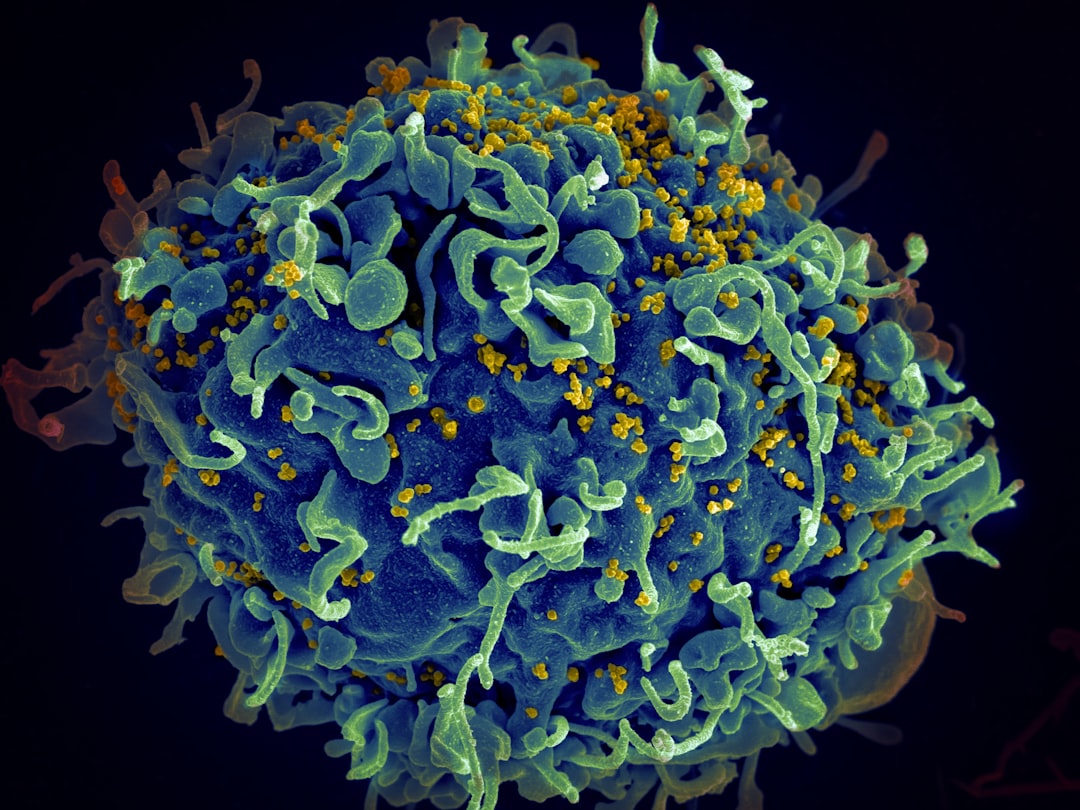
Malaria parasites are unusual, early-diverging single-celled organisms. One of their atypical features is their mode of cell division. Most cells replicate their genome, segregate the copies into two nuclei and then split the whole cell in two: a process called binary fission. Malaria parasites, by contrast, multiply primarily by schizogony. Each cell replicates its genome several times over, asynchronously, generating many nuclei within the same cytoplasm, before splitting into many new daughter cells in a single mass event. All malaria species do this, but there are stark differences between species in how long schizogony takes and how many progeny each cell can produce. These datasets were generated to compare the process of schizogony in two different species. We used GM cell lines in which we can follow de novo DNA replication at high spatial and temporal resolution, at both whole-cell and single-molecule levels. We establish the dynamics of schizogony, highlighting similarities and differences between two species and setting parameters for future studies of interventions that could interfere with parasite replication.
Featured Image

Photo by Sangharsh Lohakare on Unsplash
Why is it important?
Understanding the process of replication in malaria parasites is important because the rate of parasite replication is fundamentally important to malarial disease: humans who have a high burden of parasites in their blood are most likely to suffer severe malaria.
Perspectives
Understanding the process of replication in malaria parasites is important because the rate of parasite replication is fundamentally important to malarial disease: humans who have a high burden of parasites in their blood are most likely to suffer severe malaria.
Catherine Merrick
University of Cambridge
Read the Original
This page is a summary of: DNA replication dynamics during erythrocytic schizogony in the malaria parasites Plasmodium falciparum and Plasmodium knowlesi, PLoS Pathogens, June 2022, PLOS,
DOI: 10.1371/journal.ppat.1010595.
You can read the full text:
Contributors
The following have contributed to this page